
BIOLOGY INTERACT
ProC-Chromatin (Protein Co-localizer on Chromatin)
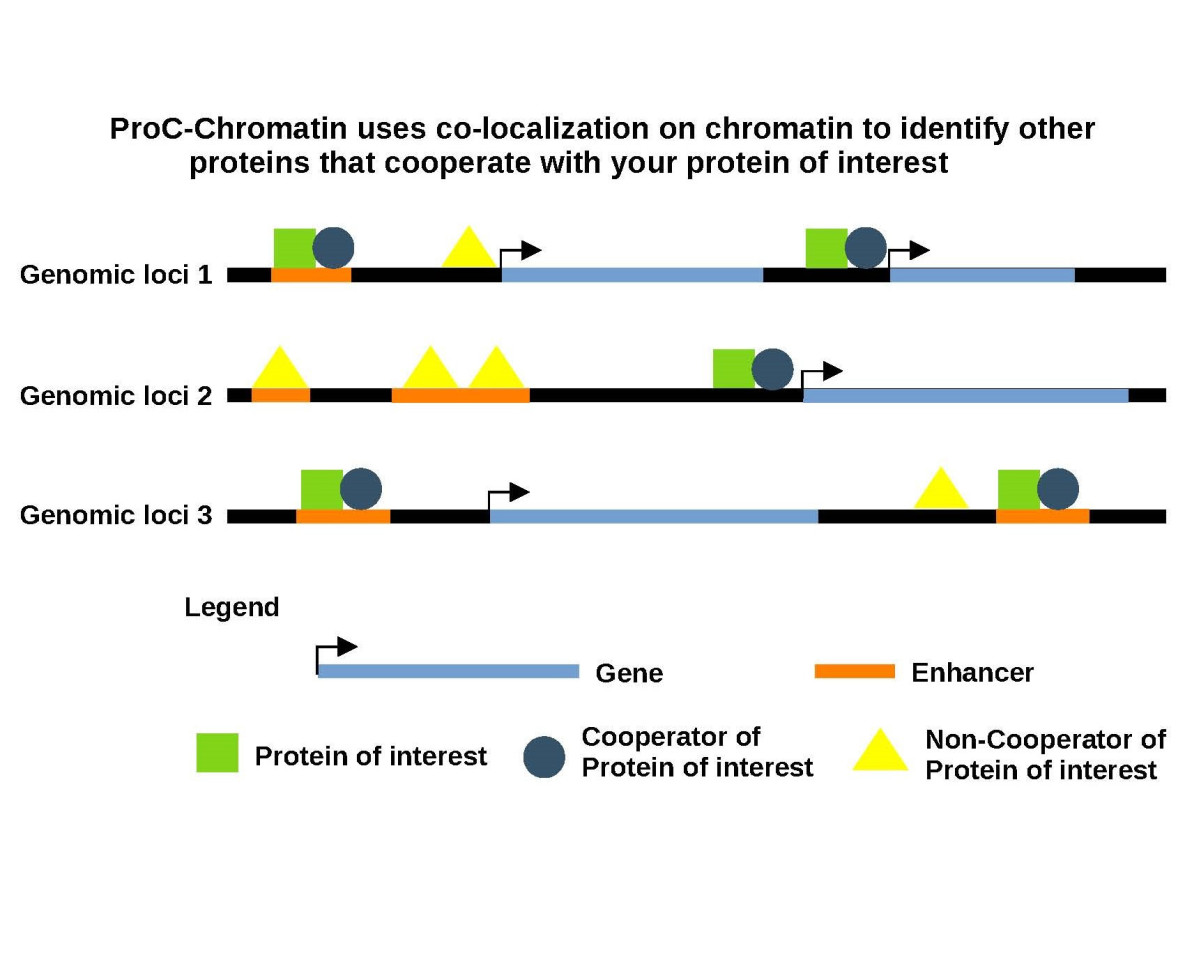
Gene expression is regulated by groups of proteins that bind at promoters or distal regulatory elements. These proteins which include general transcription factors, sequence specific transcription factors, histone modifiers and chromatin remodelers can interact with each other on chromatin directly, indirectly, or they can bind in the proximity of each other.
ChIP-seq has been used to map the localization of a large number of proteins across a diverse set of cell types and this data can be leveraged to understand how proteins cooperate with each other to regulate gene expression.
ProC-Chromatin uses statistical methods to screen hundreds of high quality ChIP-seq experiments to identify the set of proteins that most significantly co-localize with your protein of interest (POI) on chromatin.
The output of this analysis will include a list of proteins that are ranked based on their significance of colocalization with your POI along with associated statistics, as well as the DNA-binding sites where these proteins co-localize.
This analysis can help you identify other proteins which your POI may cooperate to regulate gene expression and it can help you design experiments to test their cooperation.
If you have a protein of interest that you would like to analize with ProC-Chromatin, you can provide us one of the following:
-
The genomic locations where your protein of interest binds
-
The raw ChIP-seq data for your protein of interest, or other sequencing based methods that identify association of proteins with chromatin
-
Contact us to inquire if we have already found high quality dataset that provide the genomic location where your protein of interest binds
Please contact us to inquire about how ProC-Chromatin works, how it can be tailored to your needs, as well as quotes.